Host - Parasites
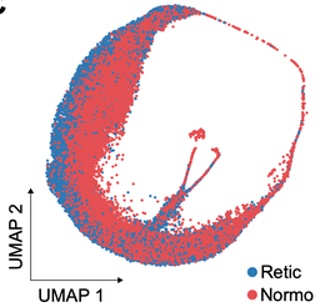
Plasmodium berghei infected red blood cell
legacy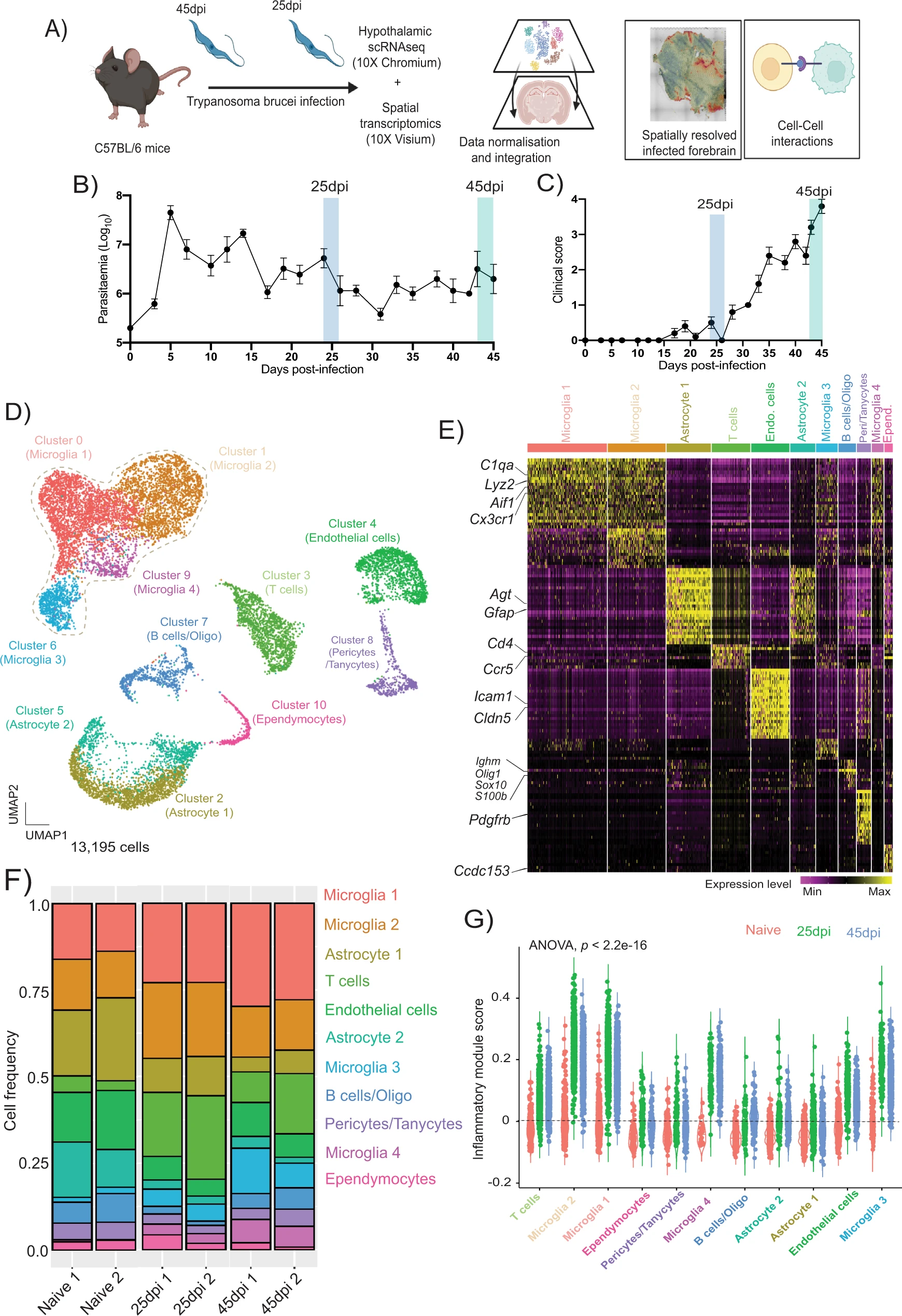
Mouse brain cell during Trypanosoma brucei infection
paraCellHuman-COVID-19
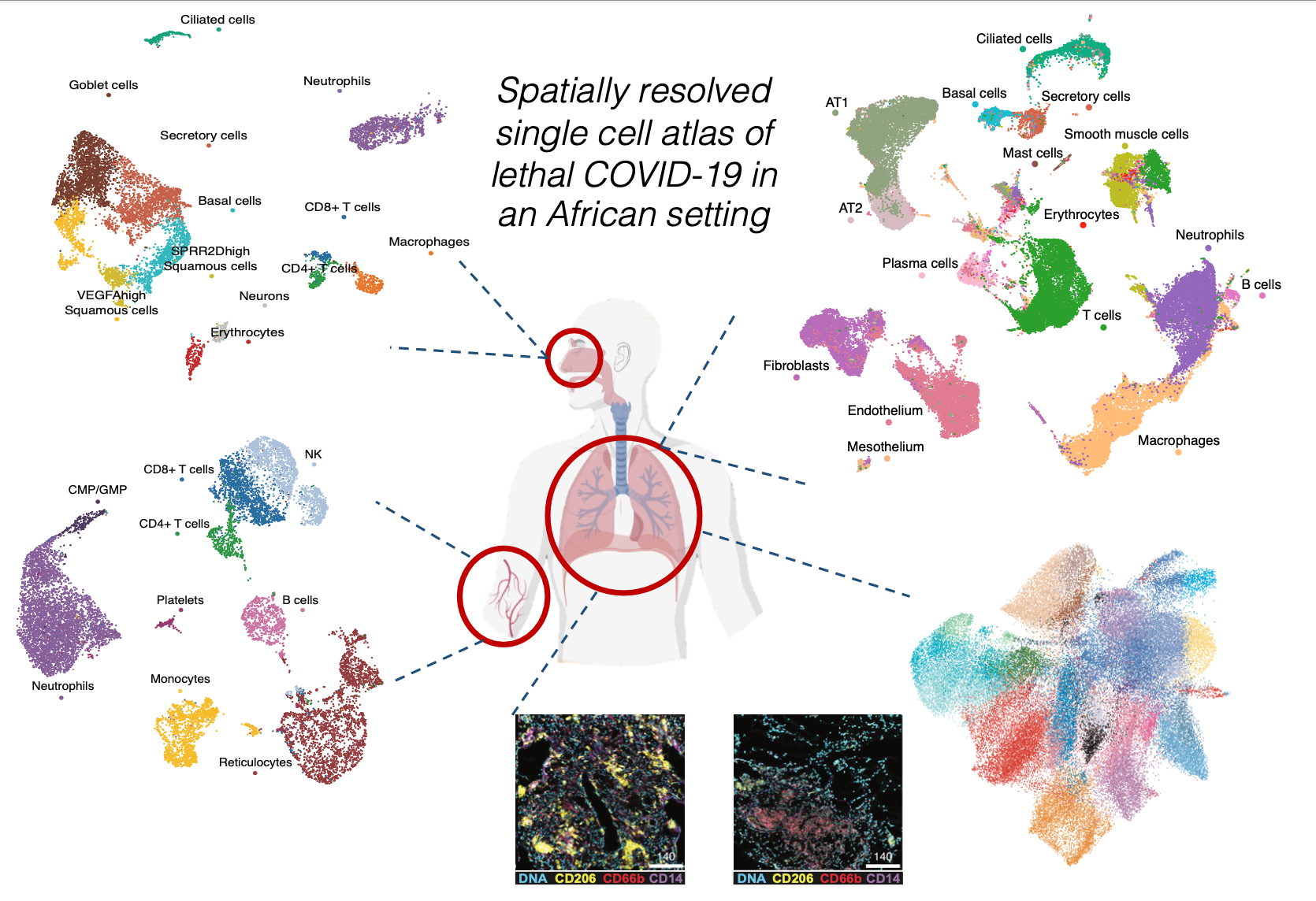
To see a table of all published data sets with paraCell atlases:Click Here
To use the full breadth of paraCell, please have a look at the tutorial on the WIKI. Helpful might be the advanced genes searches.


